Long Reads Potential in Transcriptomics
Over the last 10 years, a new third generation of sequencing (TGS) platforms, represented by Oxford Nanopore and Pacific Biosciences, have strongly emerged as novel sequencing methods. These technologies -in contrast to the traditional short-read Illumina- are able to sequence single molecules thanks to the utilization, respectively, of nanopores and microwells, and they can produce long (> tens of thousands of nucleotides) reads. These two properties – single molecule sequencing and long reads- create exciting opportunities for nucleic acid research. They not only resolve long-range phasing problems intractable to the short reads, but they also couple sequencing to the detection of nucleotide modifications such as methylation, resulting into a simultaneous measurement of the genome and the epigenome.

Applied to transcriptome research, long-read RNA sequencing (lrRNA-seq) has important advantages. Long read sequencing (LRS) can obtain full-length transcripts, making possible the accurate detection of isoforms, alleles, and transcripts from gene families, traditionally hard to resolve by
short reads. Additionally, the Nanopore technology can perform direct sequencing of RNA molecules (dRNA-seq) and capture their nucleotide modifications, enabling new venues for epitranscriptomics research. LRS have already made breakthroughs such as the completion of the human genome from telomere to telomere and is the driving technology of the Vertebrate Genome and Earth BioGenome projects. Today, LRS has replaced short reads in genome assembly, and it is generally accepted by the community that no genome sequencing project is further conceived without the utilization of LRS. We anticipate that, in the same way that LRS has conquered the sequencing of genomes, lrRNA-seq will be the preferred transcriptome profiling technology in the near future. However, lrRNA-seq is not an error-free technology. Limitations still exist at the library preparation, mapping, transcript reconstruction and quantification steps. Upcoming applications such as single-cell sequencing, or allele-specific expression require well established protocols and benchmarked analysis pipelines. Additionally, the power of LRS to dissect the regulatory biology of RNAs needs to be further explored.
LongTREC Research Objectives
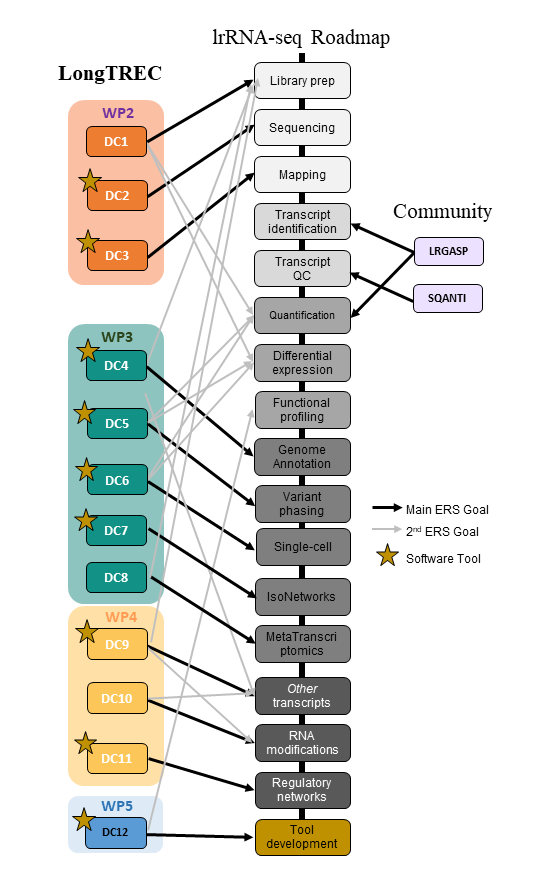
LongTREC addresses four important aspects for the consolidation of LRS as a robust and widely adopted transcriptomics technology, distributed into four work packages:
- Improve library preparation, sequencing, and pre-processing steps to obtain better lrRNA-seq data (WP2)
- Develop lrRNA-seq methods and pipelines to address pending questions in transcriptome biology (WP3).
- Advance the LRS transcriptomics field with multi-omics applications (WP4).
- Create community-wide and user-friendly tools for strong impact (WP5).
An European Doctoral Network
All the above reveals that lrRNA-seq is an effervescent field, where new experimental protocols, biological applications and analysis solutions are constantly emerging in pace with the rapid improvements in third-generation sequencing technologies. lrRNA-seq has already displayed great potential, but much work is still needed. A new generation of computational biologists is essential to consolidate current methods, develop new tools, envision new applications, and lead the unstoppable transition of transcriptome analysis to effectively become LRS-based.
In the LongTREC MSCA-DN we propose to address these challenges by collaborative research and training of 12 Doctoral candidates (DC) who will acquire the set of competences to achieve this transformation. To make this ambitious goal possible we have gathered the current European leaders of the lrRNA-seq technologies, both at experimentation and computation.
Individual research projects are structured in three Objectives that form the basis for three core chapters of PhD thesis dissertations. In most cases, these Objectives involve the development of experimental protocols, different levels of data analysis, and software implementations. All these components are extensively covered by the LongTREC doctoral training programme, which includes modules in genomic technologies, statistical sciences, and software development.
All LongTREC research projects are backed by extensive preliminary data and expertise of supervisors who count with high-impact publications in the incipient lrRNA-seq field. Additionally, secondments -in industry and academia- are carefully designed to support research projects with the expertise of our network.